-
Notifications
You must be signed in to change notification settings - Fork 4
Examples
Both examples consider the following combinations of hyperparameters used for Bayesian network learning:
- K2 metric;
- K2 metric with gaussian mixtures (GMM);
- K2 metric with GMM and logit nodes;
- K2 with initial structure.
All the examples are executed using cross-validation, the data is preproccesed as follows:
data.dropna(inplace=True)
data.reset_index(inplace=True, drop=True)
encoder = preprocessing.LabelEncoder()
discretizer = preprocessing.KBinsDiscretizer(n_bins=5, encode='ordinal', strategy='quantile')
p = pp.Preprocessor([('encoder', encoder), ('discretizer', discretizer)])
discretized_data, est = p.apply(train)The data set contains 9 variables with 442 samples. The target variable for prediction in the following example is 'Depth'. The variable is also used to visually evaluate sampling quality via distribution plot.
To sample using K2 metric the following code can be used:
train, validation = train_test_split(data, test_size=0.1)
bn = Nets.HybridBN(has_logit=False, use_mixture=False)
bn.add_nodes(info)
bn.add_edges(discretized_data, scoring_function=('K2',K2Score))
bn.fit_parameters(train)
# prediction
val_pred = bn.predict(validation.iloc[:,:8], 5)
# sampling
sample = bn.sample(5000, parall_count=5)
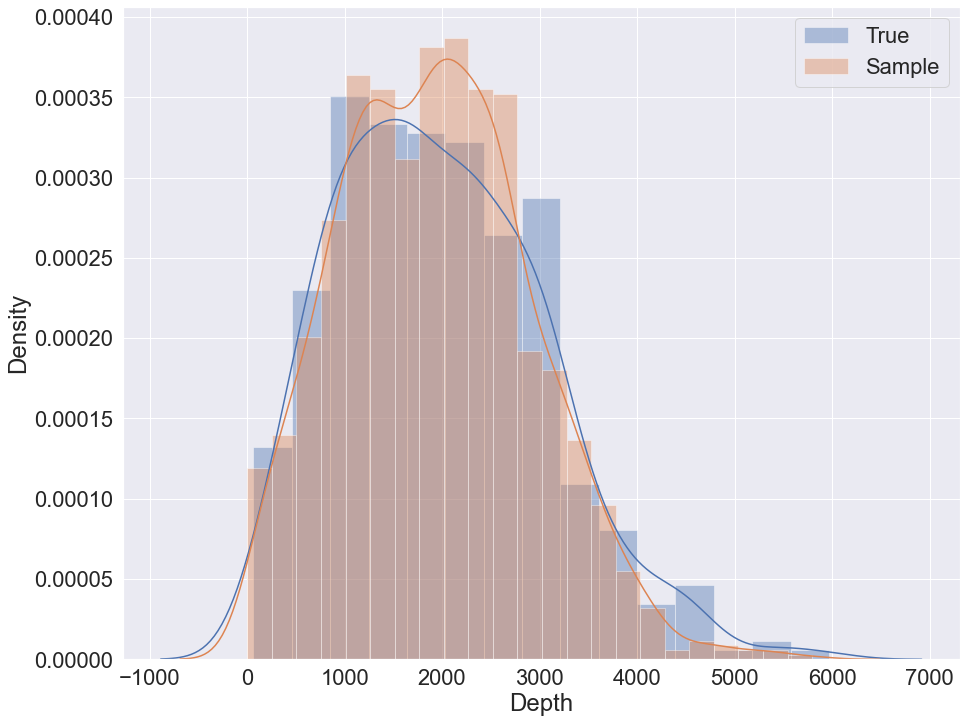
To sample using K2 with GMM the following code can be used:
train, validation = train_test_split(data, test_size=0.1)
bn = Nets.HybridBN(has_logit=False, use_mixture=True)
bn.add_nodes(info)
bn.add_edges(discretized_data, scoring_function=('K2',K2Score))
bn.fit_parameters(train)
# prediction
val_pred = bn.predict(validation.iloc[:,:8], 5)
# sampling
sample = bn.sample(5000, parall_count=5)
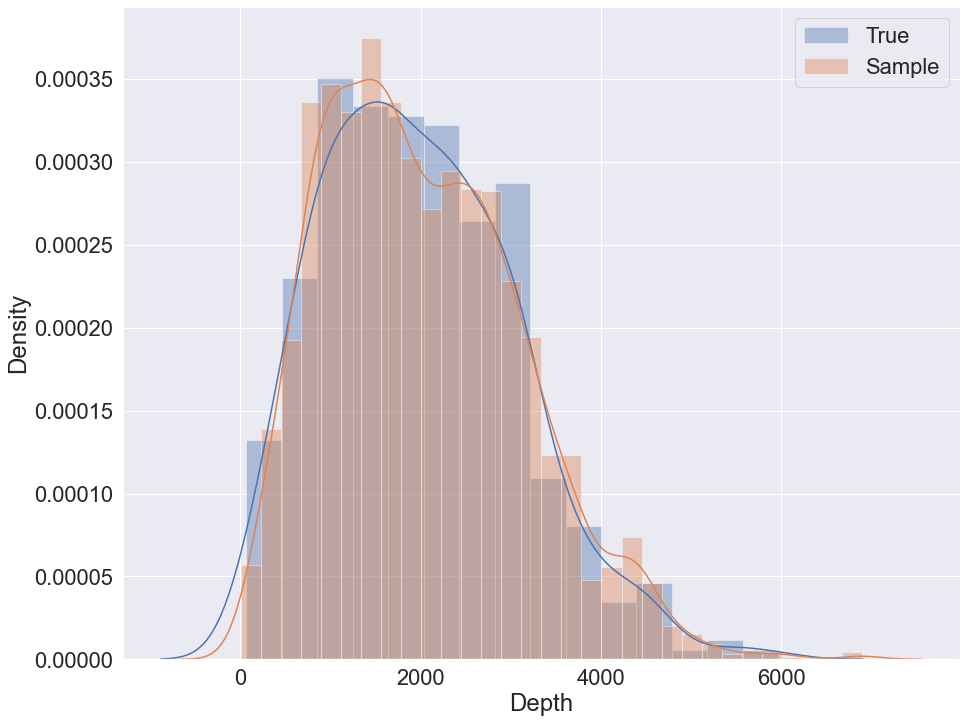
To sample using K2 with GMM and logit nodes the following code can be used:
train, validation = train_test_split(data, test_size=0.1)
bn = Nets.HybridBN(has_logit=True, use_mixture=True)
bn.add_nodes(info)
bn.add_edges(discretized_data, scoring_function=('K2',K2Score))
bn.fit_parameters(train)
# prediction
val_pred = bn.predict(validation.iloc[:,:8], 5)
# sampling
sample = bn.sample(5000, parall_count=5)
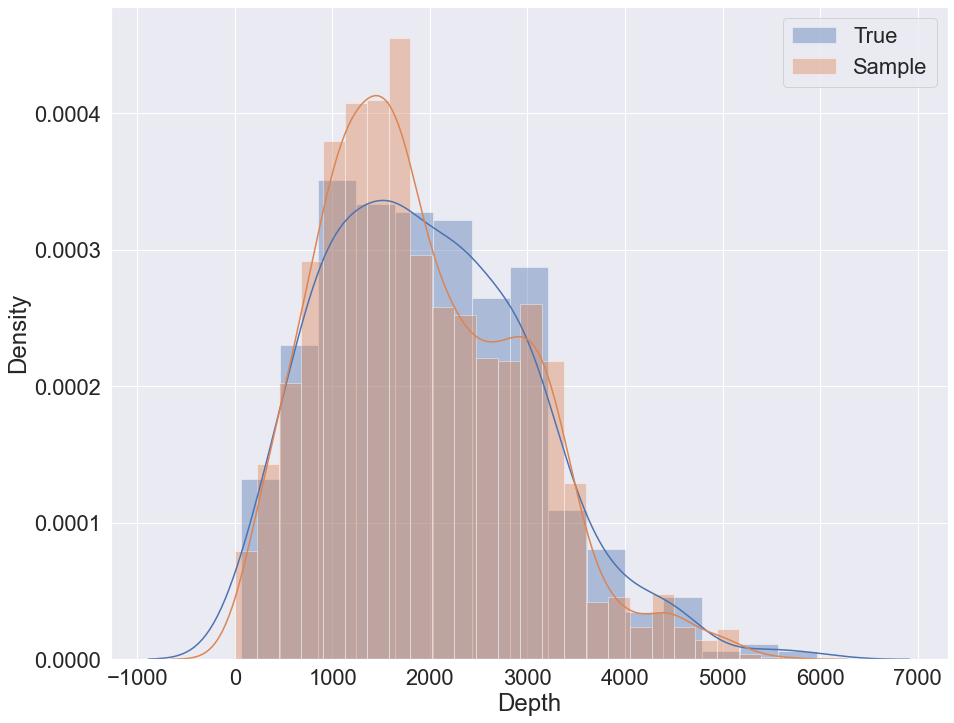
To sample using K2 and initial structure the following code can be used:
params = {'init_nodes': ['Tectonic regime', 'Period', 'Lithology', 'Structural setting', 'Gross','Netpay', 'Porosity','Permeability', 'Depth'],
'init_edges':[('Period', 'Permeability'), ('Structural setting', 'Netpay'), ('Gross', 'Permeability')],}
train, validation = train_test_split(data, test_size=0.1)
bn = Nets.HybridBN(has_logit=True, use_mixture=True)
bn.add_nodes(info)
bn.add_edges(discretized_data, scoring_function=('K2',K2Score), params=params)
bn.fit_parameters(train)
# prediction
val_pred = bn.predict(validation.iloc[:,:8], 5)
# sampling
sample = bn.sample(5000, parall_count=5)

The second example is similar to the previous one, but carried out on different data set. Social data set consists of 30000 anonymous bank records with 9 variables each, bayesian networks were learnt on a sample with 2000 records. The target variable is 'mean_tr' which is mean transaction of client.
The code used to sample from social data set is identical to the geological dataset.








| Hyperparameters combinations | Geological data MSE | Social data MSE |
|---|---|---|
| K2 | 1014.59 | 6066.5 |
| K2 + GMM | 974.35 | 5149.5 |
| K2 + GMM + logit | 1018.84 | 6657.93 |
| K2 + initial structure | 1056.06 | 12506.47 |
Each module has the following files (.py), their content has described in API docs:
- Controller (endpoints, routes, queries)
- Service (core functions)
- Schema (docs schemas)
- Models (table descriptions for database)
