-
Notifications
You must be signed in to change notification settings - Fork 16
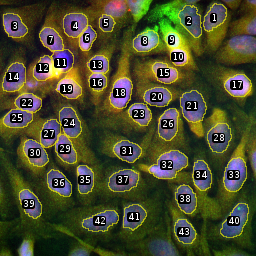
Intensity Per Nucleus Tool
The tool segments the nuclei in the dapi or hoechst channel of the image and measures the mean intensity per nuclei in the other channels of the image. The macro can be applied recursively to all images in a folder and its subfolders.
You can find the source-code in github here: intensity_per_nucleus_tool
To install the tool save the file intensity_per_nucleus_tool.ijm into the folder macros/toolsets of your FIJI installation.
Select the "intensity_per_nucleus_tool" toolset from the >> button of the ImageJ launcher.
- The first button on the toolset opens this help page.
- The m-button runs the analysis on the current image.
- The b-button runs the analysis on all the images in a given folder.
You can find example input images here:
The input image must be a multi-channel image with at least two channels. One of the channels must contain a staining of the nuclei, while the other channels contain the signals to be measured. By default the images are expected to have the .nd-file extension. A channel with the name hoechst or dapi is by default considered as the channel containing the nuclei.
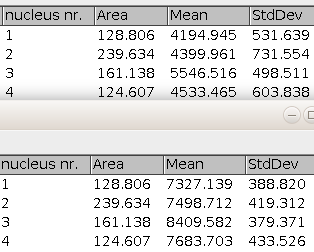
First set the features you want to measure via the ImageJ-menu Analyze>Set Measurements.
For a manual analysis, for example to test the tool, open an image and press the m-button.
For a batch analysis, press the b-button and select the root-folder of the directory-tree that contains your input images.
The method to detect the nuclei works in the following way:
- The image is scaled down to a smaller size using bilinear interpolation
- An auto-threshold is set on the image. By default the
Huangmethod is used. - The image is converted to a mask and holes in the mask are filled with the
Fill Holescommand of ImageJ - A binary watershed is applied to the mask
- The mask is scaled back to the original image size using bilinear interpolation again
- ImageJ's
particle analyzeris used to create a roi for each nucleus, to remove objects with a too small area and to remove objects touching the border of the image.
-
nuclei channels
: A comma separated list of channel-names. These are the possible names of the channel containing the nuclei. -
scale factor
: The factor by which the image is scaled down before the watershed is applied. The bigger the value the smaller the risk of getting over-segmentation from the watershed algorithm. In the same time a bigger value will provide less accurate contours of the nuclei. -
min size
: The minimum area in square-micrometer of the objects that will be considered nuclei. -
thresholding method
: The auto-threshold method used to create the mask for the segmentation of the nuclei.
-
file ext.
: The file extension of the input-image-files.

 Volker Bäcker
Volker Bäcker